Autoencoders¶
An autoencoder is a feed forward neural network which is trained to map its input to itself via the representation formed by the hidden units. The optimisation problem for input data \(\vec{x}_1,\dots,\vec{x}_N\) is stated as:
Of course, without any constraints this is a simple task as the model will just try to learn the identity. It becomes a bit more challenging when we restrict the size of the intermediate representation (i.e., the number of hidden units). An image with several hundred input points can not be squeezed in a representation of a few hidden neurons. Thus, it is assumed that this intermediate representation learns something meaningful about the problem. Of course, using this simple technique without any additional regularization only works if the number of hidden neurons is smaller than the number of dimensions of the image.
As a dataset for this tutorial, we use a subset of the MNIST dataset which needs to
be unzipped first. It can be found in examples/Supervised/data/mnist_subset.zip.
The full example program can be found in AutoEncoderTutorial.cpp.
The following includes are needed for this tutorial:
#include <shark/Data/Pgm.h> //for exporting the learned filters
#include <shark/Data/SparseData.h>//for reading in the images as sparseData/Libsvm format
#include <shark/Models/LinearModel.h>//single dense layer
#include <shark/Models/ConcatenatedModel.h>//for stacking layers
#include <shark/ObjectiveFunctions/ErrorFunction.h> //the error function for minibatch training
#include <shark/Algorithms/GradientDescent/Adam.h>// The Adam optimization algorithm
#include <shark/ObjectiveFunctions/Loss/SquaredLoss.h> // squared loss used for regression
#include <shark/ObjectiveFunctions/Regularizer.h> //L2 regulariziation
Training autoencoders¶
Training an autoencoder is straight forward in shark. We just setup two neural networks, one for encoding and one for decoding. Those are then concatenated to form one autoencoder network:
//We use a dense lienar model with rectifier activations
typedef LinearModel<RealVector, RectifierNeuron> DenseLayer;
//build encoder network
DenseLayer encoder1(inputs,hidden1);
DenseLayer encoder2(encoder1.outputShape(),hidden2);
auto encoder = encoder1 >> encoder2;
//build decoder network
DenseLayer decoder1(encoder2.outputShape(), encoder2.inputShape());
DenseLayer decoder2(encoder1.outputShape(), encoder1.inputShape());
auto decoder = decoder1 >> decoder2;
//Setup autoencoder model
auto autoencoder = encoder >> decoder;
Note that for the deeper layers we use the shape of the output of the previous layers (in this case just a 1-d shape with the number of neurons) to specify the shape of the input of the next layer.
Next, we set up the objective function. This should by now be looking quite familiar. We set up an ErrorFunction with the model and the squared loss. Here we enable minibatch training to speed up the training progress. We create the LabeledData object from the input data by setting the labels to be the same as the inputs. Finally we add two-norm regularisation by creating an instance of the TwoNormRegularizer class:
//create the objective function as a regression problem
LabeledData<RealVector,RealVector> trainSet(data.inputs(),data.inputs());//labels identical to inputs
SquaredLoss<RealVector> loss;
ErrorFunction<> error(trainSet, &autoencoder, &loss, true);//we enable minibatch learning
TwoNormRegularizer<> regularizer(error.numberOfVariables());
error.setRegularizer(regularisation,®ularizer);
initRandomNormal(autoencoder,0.01);
Lastly, we optimize the objective using Adam:
Adam<> optimizer;
error.init();
optimizer.init(error);
std::cout<<"Optimizing model "<<std::endl;
for(std::size_t i = 0; i != iterations; ++i){
optimizer.step(error);
if(i % 100 == 0)
std::cout<<i<<" "<<optimizer.solution().value<<std::endl;
}
autoencoder.setParameterVector(optimizer.solution().point);
Visualizing the autoencoder¶
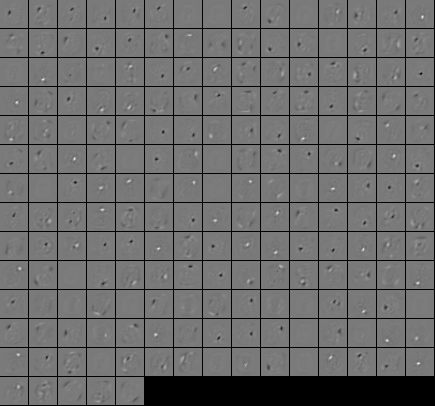
After training the different architectures, we printed the feature maps of the first layer (i.e., the input weights of the hidden neurons ordered according to the pixels they are connected to). Let’s have a look.
exportFiltersToPGMGrid(“features”,encoder1.matrix(),28,28);
What next?¶
The next tutorials covers Variational Autoencoders

